
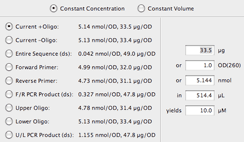

This application also supports different units for concentration and volume.Īll these analyses can be done with a single free online tool that includes a direct link to the respective oligo order menu. Constant b adopts the value of 4 for non-self-complementary sequences or. The appropriate oligo synthesis scale can also be decipered.ĭetermines how much volume is needed to dilute a highly concentraged oligo stock solution to a specific lower concentration. Finally, a list of guidelines to calculate the Tm of short DNA sequences with a. Tests your oligo against itself or another oligo for dimer formation and performs a PCR viability check for a primer pairĬalculates the optical density (OD), mass, amount and molecular weight (MW) of your oligo for specific experimental conditions. For deeper analysis of your sequence, please check out our Codon Optimization tool, or to compare two sequences at the DNA or protein level. The sequence should begin with the start codon (ATG) and be in a multiple of 3 for a complete codon sequence.

The entire IUPAC DNA alphabet is supported, and the case of each input sequence character is maintained. Input your DNA sequence below to retrieve the translated amino acid sequence. Mfold is an adaptation of the mfold package by Zuker and Jaeger which was modified to work with the Wisconsin Package.Īligns your sequence against human, mouse and rat Refseq database entries and provides a comprehensive BLASTn report. Reverse Complement converts a DNA sequence into its reverse, complement, or reverse-complement counterpart. Predict the secondary structure pf DNA and RNA oligos using a mfold and RNAfold program. Type or paste your DNA sequence below and automatically retrieve the reverse, complement, or reverse complement sequence. The entire IUPAC DNA alphabet is supported. Check to make sure your ligase is active. Reverse Complement converts a DNA sequence into its reverse, complement, or reverse-complement counterpart. Visualize your ligation reactions on a gel. TM calculatorĭetermines all physical properties such as GC content, melting temperature (Tm), molecular weight (MW), extinction coefficient (EC) and complement sequences In this article, we share seven must-have tips for your ligation reactions: Consider your cloning strategy. MOPS allows you to analyse and calculate any physical property of your DNA or RNA oligo sequence: Our multifunctional oligo property analysis tool (MOPS) is an oligo calculator inclusive a dilution calculator, a TM calculator and much more! The one-stop solution for quick and easy oligo analysis


 0 kommentar(er)
0 kommentar(er)
